| This post was kindly contributed by The DO Loop - go there to comment and to read the full post. |
This article describes best practices and techniques that every data analyst should know before bootstrapping in SAS.
The bootstrap method is a powerful statistical technique, but it can be a challenge to implement it efficiently.
An inefficient bootstrap program can take hours to run, whereas a well-written program can give you an answer in an instant.
If you prefer “instants” to “hours,” this article is for you! I’ve compiled dozens of resources that explain how to compute bootstrap statistics in SAS.
Overview: What is the bootstrap method?
Recall that a bootstrap analysis enables you to investigate the sampling variability of a statistic without making any distributional assumptions about the population. For example, if you compute the skewness of a univariate sample, you get an estimate for the skewness of the population. You might want to know the range of skewness values that you might observe from a second sample (of the same size) from the population. If the range is large, the original estimate is imprecise. If the range is small, the original estimate is precise. Bootstrapping enables you to estimate the range by using only the observed data.
In general, the basic bootstrap method consists of four steps:
- Compute a statistic for the original data.
- Use the DATA step or PROC SURVEYSELECT to resample (with replacement) B times from the data. The resampling process should respect the null hypothesis or reflect the original sampling scheme. For efficiency, you should
put all B random bootstrap samples into a single data set. -
Use BY-group processing to compute the statistic of interest on each bootstrap sample. The BY-group approach is much faster than using macro loops.
The union of the statistic is the bootstrap distribution, which approximates the sampling distribution of the statistic under the null hypothesis. Don’t forget to turn off ODS when you run BY-group processing! - Use the bootstrap distribution to obtain estimates for the bias and standard error of the statistic and confidence intervals for parameters.
The links in the previous list provide examples of best practices for bootstrapping in SAS. In particular, do not fall into the trap of using a macro loop to “resample, analyze, and append.” You will eventually get the correct bootstrap estimates, but you might wait a long time to get them!
The remainder of this article is organized by the three ways to perform bootstrapping in SAS:
- Programming: You can write a SAS DATA step program or a SAS/IML program that resamples from the data and analyzes each (re)sample. The programming approach gives you complete control over all aspects of the bootstrap analysis.
- Macros: You can use the %BOOT and %BOOTCI macros that are supplied by SAS. The macros handle a wide variety of common bootstrap analyses.
- Procedures: You can use bootstrap options that are built into several SAS procedures. The procedure internally implements the bootstrap method for a particular set of statistics.
Programming the basic bootstrap in SAS
The articles in this section describe how to program the bootstrap method in SAS for basic univariate analyses, for regression analyses, and for related resampling techniques such as the jackknife and permutation tests. This section also links to articles that describe how to generate bootstrap samples in SAS.
Examples of basic bootstrap analyses in SAS
- The basic bootstrap in SAS:
SAS enables you to resample the data by using PROC SURVEYSELECT. When coupled with BY-group processing, you can perform a very efficient bootstrap analysis in SAS, including the estimate of standard errors and percentile-based confidence intervals. - The basic bootstrap in SAS/IML: The SAS/IML language provides a compact language for bootstrapping, as shown in this basic bootstrap example.
- The smooth bootstrap: As originally conceived,
a bootstrap sample contains replicates of the data. However, there are situations when “jittering” the data provides a better approximation of the sampling distribution. - Bias-corrected and adjusted (BCa) confidence interval: For highly skewed data, the percentile-based confidence intervals are less efficient than the BCa confidence interval.
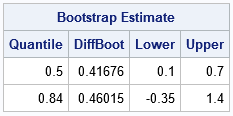
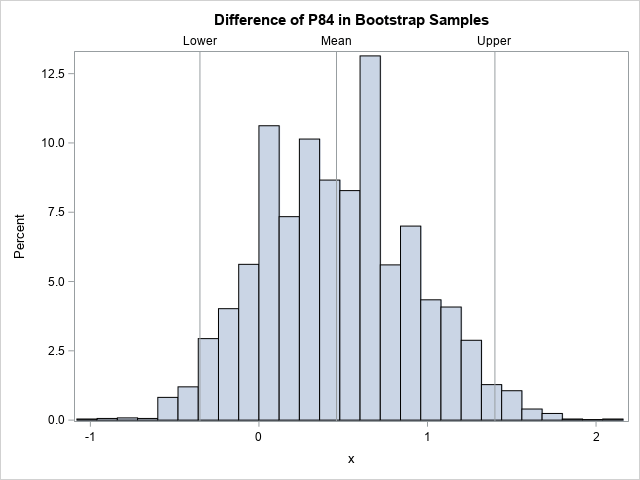
- Bootstrap the difference of means between two groups: This example shows how to bootstrap a statistic in a two-sample t test.
-
Compute a p-value for a bootstrap analysis.
Examples of bootstrapping for regression statistics
When you bootstrap regression statistics, you have two choices for generating the bootstrap samples:
- Case resampling:
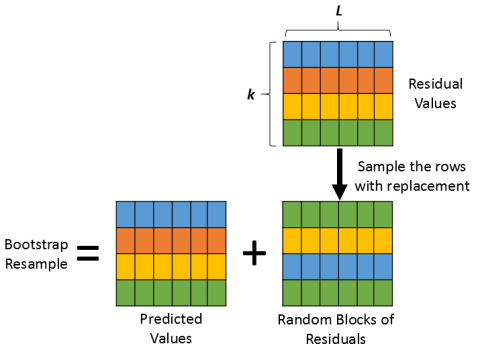
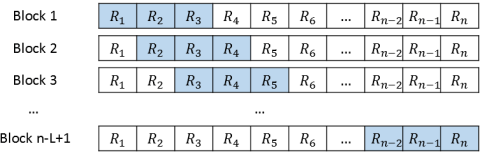
You can resample the observations (cases) to obtain bootstrap samples of the responses and the explanatory variables. - Residual resampling:
Alternatively, you can bootstrap regression parameters by fitting a model and resampling from the residuals to obtain new responses.
Jackknife and permutation tests in SAS
- The jackknife method: The jackknife in an alternative nonparametric method for obtaining standard errors for statistics. It is deterministic because it uses leave-one-out samples rather than random samples.
-
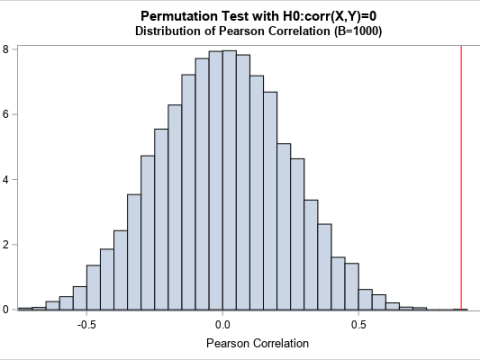
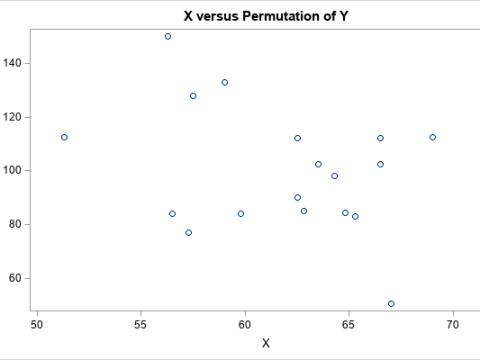
Permutation tests: A
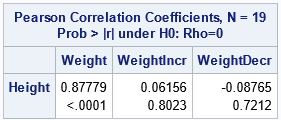
permutation test is a resampling technique that is closely related to the bootstrap. You permute the observations between two groups to test whether the groups are significantly different.
Generate bootstrap sampling
An important part of a bootstrapping is generating multiple bootstrap samples from the data. In SAS, there are many ways to obtain the bootstrap samples:
- Sample with replacement: The most common resampling technique is to randomly sample with replacement from the data. You can use the SAS DATA step, the SURVEYSELECT procedure, or the SAMPLE function in SAS/IML.
-
Samples in random order:
It is sometimes useful to generate random samples in which the order of the observations is randomly permuted. -
Balanced bootstrap resampling:
Instead of random samples, some experts advocate a resampling algorithm in which each observation appears exactly B times in the union of the B bootstrap samples.
Bootstrap macros in SAS
The SAS-supplied macros %BOOT, %JACK, and %BOOTCI,
can perform basic bootstrap analyses and jackknife analyses. However, they require a familiarity with writing and using SAS macros. If you are interested, I wrote an example that shows how to use the %BOOT and %BOOTCI macros for bootstrapping. The documentation also provides several examples.
SAS procedures that support bootstrapping
Many SAS procedures not only compute statistics but also provide standard errors or confidence intervals that enable you to infer whether an estimate is precise. Many confidence intervals are based on distributional assumptions about the population. (“If the errors are normally distributed, then….”) However, the following SAS procedures provide an easy way to obtain a distribution-free confidence interval by using the bootstrap. See the SAS/STAT documentation for the syntax for each procedure.
- PROC CAUSALMED introduced the BOOTSTRAP statement in SAS/STAT 14.3 (SAS 9.4M5). The statement enables you to compute bootstrap estimates of standard errors and confidence intervals for various effects and percentages of total effects.
- PROC MULTTEST supports the BOOTSTRAP and PERMUTATION options, which enable you to compute estimates of p-values that make no distributional assumptions.
- PROC NLIN supports the BOOTSTRAP statement, which computes bootstrap confidence intervals for parameters and bootstrap estimates of the covariance of the parameter estimates.
- PROC QUANTREG supports the CI=RESAMPLING option to construct confidence intervals for regression quantiles.
-
The SURVEYMEANS, SURVEYREG, SURVEYLOGISTIC, SURVEYPHREG, SURVEYIMPUTE and SURVEYFREQ procedures introduced the VARMETHOD=BOOTSTRAP option SAS 9.4M5. The option enables you to compute bootstrap estimates of variance.
With the exception of SURVEYIMPUTE, these procedures also support jackknife estimates. The jackknife is similar to the bootstrap but uses a leave-one-out deterministic scheme rather than random resampling. - PROC TTEST introduced the BOOTSTRAP statement in SAS/STAT 14.3. The statement enables you to compute bootstrap standard error, bias estimates, and confidence limits for means and standard deviations in t tests. In SAS/STAT 15.1 (SAS 9.4M6), the TTEST procedure provides extensive graphics that visualize the bootstrap distribution.
Summary
Resampling techniques such as bootstrap methods and permutation tests are widely used by modern data analysts. But how you implement these techniques can make a huge difference between getting the results in a few seconds versus a few hours.
This article summarizes and consolidates many previous articles that demonstrate how to perform an efficient bootstrap analysis in SAS. Bootstrapping enable you to investigate the sampling variability of a statistic without making any distributional assumptions. In particular, the bootstrap is often used to estimate standard errors and confidence intervals for parameters.
Further Reading
- Cassell, David L. (2010) “BootstrapMania!: Re-Sampling the SAS Way”, Proceedings of
the SAS Global Forum 2010 Conference. - Vickery, John (2015) “Permit Me to Permute: A Basic Introduction to Permutation Tests”,
Proceedings of
the SAS Global Forum 2015 Conference. - Wicklin, Rick (2013), “Resampling and Bootstrap Methods,” Chapter 15 in Simulating Data with SAS, SAS Institute Inc., Cary NC.
The post The essential guide to bootstrapping in SAS appeared first on The DO Loop.
| This post was kindly contributed by The DO Loop - go there to comment and to read the full post. |